Skeletonized models are available as coordinate files in PDB format (see below). Movies of 3D tissue structures are provided in our YouTube channel.
Viewer
We recommend using the PC software 'MCTrace' for viewing the wire models of tissue microstructure. The MCTrace can be downloaded from the
MCTrace page
at GitHub.
All files are plain text files. Most of the lines are based on the PDB format, a format for describing three-dimensional models of macromolecules. Therefore, PDB viewers can be used to display the model coordinates. However, the 'REMARK RM' lines for display control can be recognized only by MCTrace.
Three-dimensional coordinates
The first half of the file lines are HETATM entries. Each HETATM entry correspond to a 'node' of a wire model. The wire model is displayed by connecting these nodes. An HETATM entry consists of HETATM header, entry number, node identifier, constituent type, constituent identifier, xyz coordinates (in micrometers), density, and radius. The entry name HETATM is derived from 'hetero atom' of the original PDB format.
Connection lines
In the last half of the file, CONECT entries describe connections between node entries. Some PDB viewers including the program MCTrace can recognize the CONECT entries and display connections.
Constituent types
The three-dimensional tissue structure has many types of constituents. The constituent types of the human brain structure are: PYR, pyramidal neuron; STE, stellate neuron; ASY, aspiny stellate neuron; NEU, neuron; GLI, glial cell; UND, orphan neurites; CEL, uncategorized cell; and VES, blood vessels. The constituent type of other structures represents group names, which are determined on the basis of the three-dimensional structure.
Our micro-CT or nano-CT analysis is performed by recording x-ray images while rotating the sample. Then its 3D structure is reconstructed with the convolution back-projection method. This calculation is repeated for each tomographic slice, giving the 3D structure consisted of a large number of slices. Therefore, one 3D structure consists of thousands of images, usually in the 8-bit TIFF format. They are too large for the internet transfer, and it's impossible to place all the files online. So we provide them in DVD-ROMs if you would like. Please email mizutanilaboratory(at)gmail.com to request DVDs. They are provided at cost.
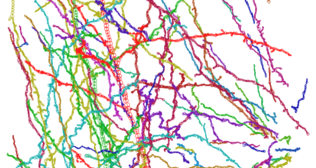
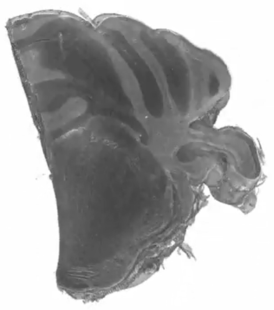

 Three-dimensional rendering of absorption coefficients.
Three-dimensional rendering of absorption coefficients. Skeletonized model.
Skeletonized model.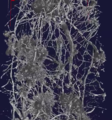 Three-dimensional rendering of absorption coefficients.
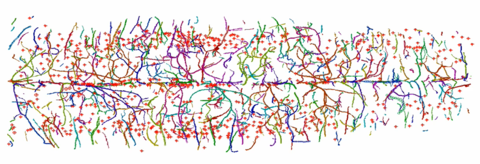
Three-dimensional rendering of absorption coefficients. Skeletonized structures of neurons and blood vessels in the inner pyramidal and inner granular layers.
Skeletonized structures of neurons and blood vessels in the inner pyramidal and inner granular layers. Three-dimensional rendering of juvenile zebra fish.
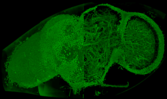
Three-dimensional rendering of juvenile zebra fish. Adult Drosophila melanogaster fruitfly.
Adult Drosophila melanogaster fruitfly.